New Binding Mode of SLURP Protein to a7 Nicotinic Acetylcholine Receptor Revealed by Computer Simulations
DOI:
https://doi.org/10.14529/jsfi180407Abstract
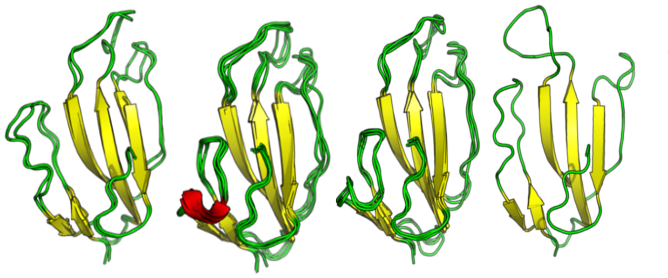
SLURP-1 is a member of three-finger toxin-like proteins. Their characteristic feature is a set of three beta strands extruding from hydrophobic core stabilized by disulfide bonds. Each beta-strand carries a flexible loop, which is responsible for recognition. SLURP-1 was recently shown to act as an endogenous growth regulator of keratinocytes and tumor suppressor by reducing cell migration and invasion by antagonizing the pro-malignant effects of nicotine. This effect is achieved through allosteric interaction with alpha7 nicotinic acetylcholine receptors (alpha-7 nAChRs) in an antagonist-like manner. Moreover, this interaction is unaffected by several well-known agents specifically alpha-bungarotoxin.
In this work, we carry out the conformational analysis of the SLURP-1 by a microsecond-long full-atom explicit solvent molecular dynamics simulations followed by clustering, to identify representative states. To achieve this timescale we employed a GPU-accelerated version of GROMACS modeling package. To avoid human bias in clustering we used a non-parametric clustering algorithm Affinity Propagation adapted for biomolecules and HPC environments. Then, we applied protein-protein molecular docking of the ten most massive clusters to alpha7-nAChRs in order to test if structural variability can affect binding. Docking simulations revealed the unusual binding mode of one of the minor SLURP-1 conformations.
References
Sadovnichy, V., Tikhonravov, A., Voevodin, V., Opanasenko, V.: “Lomonosov”: Supercomputing at Moscow State University. In: Contemporary High Performance Computing: From Petascale toward Exascale. pp. 283–307. Chapman & Hall/CRC Computational Science, Boca Raton, United States, Boca Raton, United States (2013)
Schr¨odinger, LLC: The pymol molecular graphics system, version 1.8 (2015)
Pierce, B.G., Wiehe, K., Hwang, H., Kim, B.H., Vreven, T., Weng, Z.: ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30(12), 1771–1773 (2014), DOI: 10.1093/bioinformatics/btu097
Kabsch, W., Sander, C.: Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22(12), 2577–2637 (1983), DOI: 10.1002/bip.360221211
Reshetnikov, R.V., Stolyarova, A.V., Zalevsky, A.O., Panteleev, D.Y., Pavlova, G.V., Klinov, D.V., Golovin, A.V., Protopopova, A.D.: A coarse-grained model for dna origami. Nucleic Acids Research 46(3), 1102–1112 (2017), DOI: 10.1093/nar/gkx1262
Lindorff-Larsen, K., Piana, S., Palmo, K., Maragakis, P., Klepeis, J.L., Dror, R.O., Shaw, D.E.: Improved side-chain torsion potentials for the amber ff99sb protein force field. Proteins: Structure, Function, and Bioinformatics pp. NA–NA (2010), DOI: 10.1002/prot.22711
Abraham, M.J., Murtola, T., Schulz, R., P´all, S., Smith, J.C., Hess, B., Lindahl, E.: GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015), DOI: 10.1016/j.softx.2015.06.001
Durek, T., Shelukhina, I.V., Tae, H.S., Thongyoo, P., Spirova, E.N., Kudryavtsev, D.S., Kasheverov, I.E., Faure, G., Corringer, P.J., Craik, D.J., Adams, D.J., Tsetlin, V.I.: Interaction of synthetic human slurp-1 with the nicotinic acetylcholine receptors. Scientific Reports 7(1) (2017), DOI: 10.1038/s41598-017-16809-0
Joosten, R.P., te Beek, T.A.H., Krieger, E., Hekkelman, M.L., Hooft, R.W.W., Schneider, R., Sander, C., Vriend, G.: A series of pdb related databases for everyday needs. Nucleic Acids Research 39(Database), D411–D419 (2010), DOI: 10.1093/nar/gkq1105
Lyukmanova, E.N., Shulepko, M.A., Buldakova, S.L., Kasheverov, I.E., Shenkarev, Z.O., Reshetnikov, R.V., Filkin, S.Y., Kudryavtsev, D.S., Ojomoko, L.O., Kryukova, E.V., Dolgikh, D.A., Kirpichnikov, M.P., Bregestovski, P.D., Tsetlin, V.I.: Watersoluble lynx1 residues important for interaction with muscle-type and/or neuronal nicotinic receptors. Journal of Biological Chemistry 288(22), 15888–15899 (2013), DOI: 10.1074/jbc.m112.436576
Downloads
Published
How to Cite
Issue
License
Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution-Non Commercial 3.0 License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
